Background
Entropix is a young biotech company specialising in the development of high-performance enzymes – Nature’s catalysts.
The company was founded by Dr Rob Rule and Prof David Hornby and recently expanded to new facilities in Runcorn, UK.
Entropix strives to improve the performance of naturally occurring enzymes using a process known as directed evolution. In this approach, numerous variants of the naturally occurring enzyme are created and the variants are screened according to the required performance criteria. The Entropix team has developed a technology platform combining computational and experimental skills that speeds up the evolution of resilient and high-performance enzymes tailored to fulfil their customer needs.
The enzymes generated by Entropix may be employed in a wide range of applications, including breaking down non-recyclable plastic waste and for enhanced diagnostic testing.
Challenge
There are many challenges when attempting to re-engineer the performance of an enzyme in the laboratory in a matter of a few weeks – a process that takes thousands of years of evolution in Nature. One way to compress these timescales is to design the starting gene sequence for the target enzyme using bioinformatic methods. Another is to automate the process of screening and sequencing the many variants that are generated in a typical round of direct evolution.
Entropix are often required to develop enzyme resilience under a wide range of chemical and environmental conditions. This requires the development of high-throughput selection and testing protocols followed by DNA sequencing (which can be time-consuming and costly). Entropix has recently developed efficient and cost-effective Next-Generation Sequencing methods for the analysis of enzyme variants – this is an essential step in linking DNA sequence to enzyme performance.
Solution
The challenges faced by Entropix have been significantly advanced through the collaboration with the VEC, who investigated how AI machine learning and classification models could inform the Entropix scientists ahead of testing.
This enhanced starting point (referred to as the ‘ground-state”) should lead to less screening, enabling them to shortlist mutants and lower the number of samples needed, which would be more cost-effective and reduce time spent during this process.
The VEC tested several options using the DNA sequence as an input, predicting if an enzyme would be active or inactive under specific conditions, including temperature, as part of the controlled testing environment. The results found the computer model could predict with 90%+ accuracy whether a mutated enzyme would prove effective in streamlining this process.
Entropix provided the VEC with access to large data sets that show which enzymes are typically active and inactive through DNA strings. The VEC team trained an AI system to recognise patterns using 80% of the data set. The system reserved the remaining 20% of data for testing to evaluate the accuracy of its predictions. This step was crucial as the system had no prior exposure to this particular data. Using the test data, the model achieved a 99/100 accuracy rating, which could significantly support the Entropix testing procedures in the future.
During this investigative work, the VEC team trained a range of machine learning and deep learning models to find the most accurate and valuable findings. The teams also investigated approaching the concerns as a regression problem instead of a classification problem: instead of asking: ‘Is this enzyme active or inactive?’, they asked, ‘Under what conditions will this enzyme remain active?’
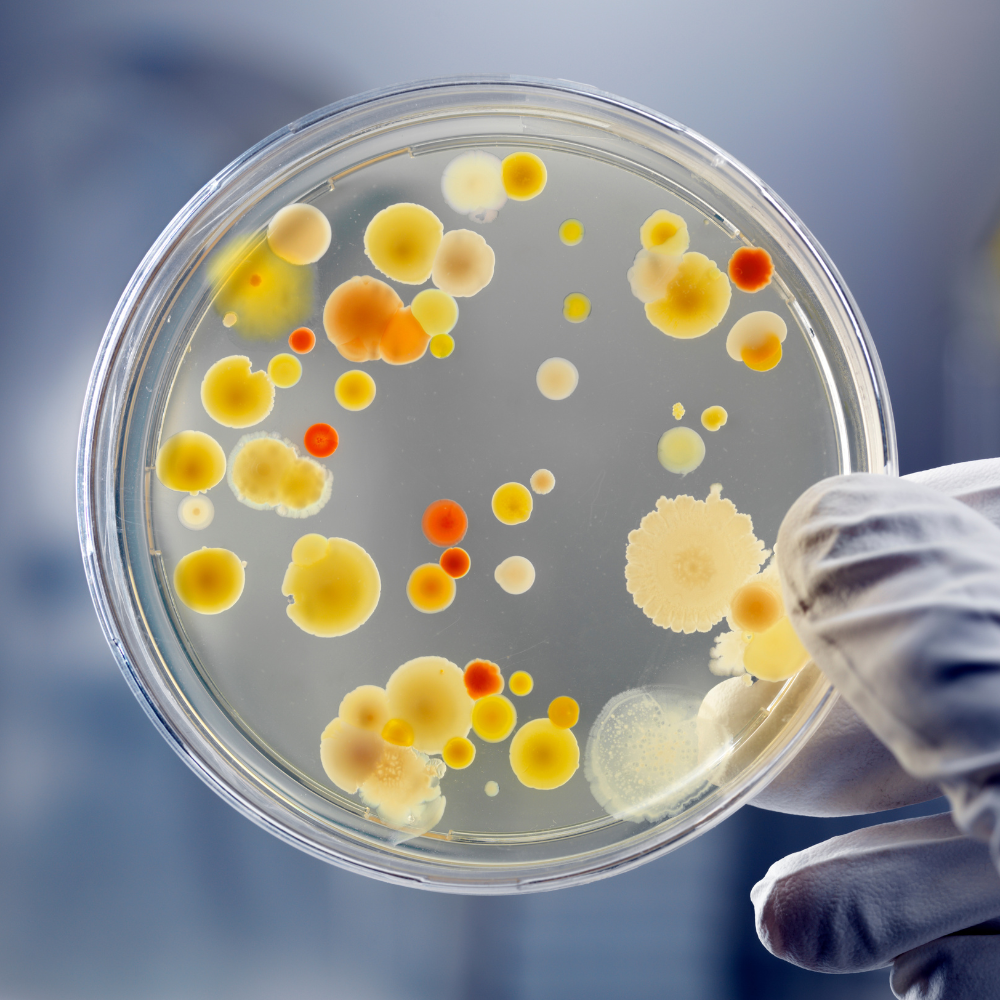
In a parallel program of work, the VEC team also explored how computerised image capture and analysis techniques could support the automation of manual testing processes. The teams developed and tested multiple processes for automatically counting and labelling bacterial colonies, producing specific classes of enzyme samples in individual petri dishes and overcoming lighting and image quality challenges.
The computer vision techniques supported the teams in better assessing the halos (tiny circles around the colonies). The size of each halo indicates the level of activity of the enzyme expressed by an individual colony. The faint halos can often be very difficult to spot with the naked eye, using different lights for identifying hard-to-see results. For this purpose, the VEC also provided Entropix with a lightbox, reducing the power of concentrated light and diffusing this to improve the accuracy of the computational image analysis results.
The testing methods developed by the VEC can automatically create digital reports, visually demonstrating the results within 3D graphs and additional charts. This numerical analysis can be used to categorise the activity of enzymes whilst quickly identifying trends and themes within the data.
Impact
Following this support, Entropix hired a new team member (who recently graduated with a PhD in biological aspects of Machine Learning) from Aston University to work alongside their molecular biologists and take forward this computational approach. This was a key step in growing the capability of the company, originally consisting of a team of three individuals, including the two founders. Entropix has significantly expanded over the past few years with no sign of slowing down.
Additionally, the VEC has supported Entropix with a competitive funding application for a UKRI grant opportunity worth around £250k, which they were successfully awarded and will play a significant role in their future investment decisions, including onboarding of machine learning capabilities.
We have had a positive and successful collaboration with the VEC over the last two years. Their skills and expertise in machine learning and image analysis have enhanced our processes in Entropix and helped us to accelerate our enzyme development activities.
Rob Rule, CEO, Entropix Ltd